DNA Shuffling.
The process of DNA exchange between different chromosomes and even between different cells is very common in Nature. This exchange of genetic material rapidly increases the diversity of a population, and is so widespread that it should be ranked as one of the most important mechanisms in evolution. It is commonly found that the DNA that is exchanged between two chomosomes or organisms is highly similar, that is to say that the strands that interchange have very similar nucleotide sequences. This high sequence similarity is important, since at some point in the exchange process the two DNA double helices must enter in contact with each other, and only the DNA strands that show high similarity will form sufficiently stable complexes.
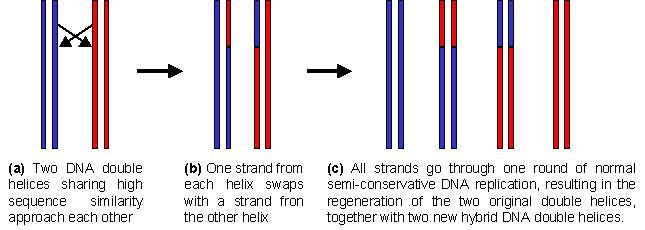
How can this process be simulated in the laboratory? Once the different DNA molecules which are to be shuffled have been isolated (step 6a in the scheme shown below), then the first step in the process is a random fragmentation of a mixture of this DNA. This is done by digestion using an enzyme called DNase, which breaks the DNA at random positions to give a collection of smaller DNA molecules of variable length (step 6b). Since the original DNA molecules from the random mutant library will all be very similar (since each differs from the others at the most only a few nucleotides at a limited number of positions), then the fragmented DNA will also be highly similar. This means that overlapping fragments will be very common and so one fragment may hybridize with another.

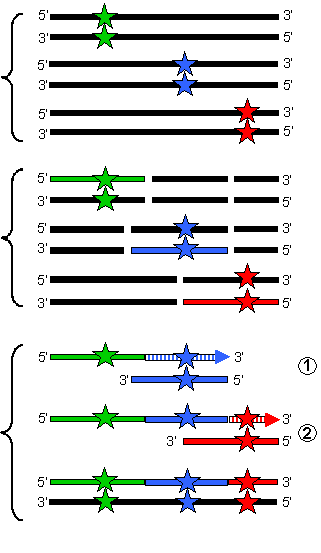
This high degree of similarity between the DNA fragments is the key to the DNA shuffling process. In successive rounds of dissociation and annealing (step 6c), homologous fragments which are derived from different DNA molecules may prime the extension of complementary DNA strands, thereby combining mutants which were originally on different DNA duplexes. The result is a collection of DNA duplexes in which all possible combinations of the original mutations are present. This ensemble of DNA molecules is called a "combinatorial library", and may be screened in and the same way as the random mutant library generated by error-prone PCR.