Directed Evolution.
Molecular biology techniques allow the manipulation of nucleotide sequence of DNA, which opens the possibility of altering the amino acid sequence, and therefore the biological functions of proteins. Directed evolution is a recent and powerful addition to the protein engineers' toolkit. By imitating the random mutagenesis of DNA and selection of desired traits that underlie the process of Darwinian natural selection, chosen proteins may be rapidly adapted to work in new and often unconventional ways. The key to success for any directed evolution project is the availability of a system to produce the target protein in a foreign cell. This requires a preliminary step that involves cloning the gene which encodes the protein of interest, and setting up a protein expression system (step 1 in the diagram shown below). The choice of host cell will be determined by the activity of the target protein. Ideally, the host cell should not naturally produce a protein that has the same activity as the target protein, since the activity of the endogenous protein may mask the detection of changes that occur in the target protein.
Once the expression system is up and running, the next step is to introduce random mutations into the DNA sequence of the gene that encodes the target protein. This may be by the use of a technique called "error-prone PCR" (step 2), which is a variation on the well established polymerase chain reaction (PCR) technique that is widely used in most molecular biology laboratories. As a consequence of the doubling in the number of copies of the gene during each cycle of PCR, one all the amazing properties of this reaction is the capacity produce billions of copies of a chosen DNA sequence in just a few hours. Using error-prone PCR, billions of copies of the gene of interest may be produced, each of which has a different set of mutations in the DNA sequence - and all in a single mornings work!
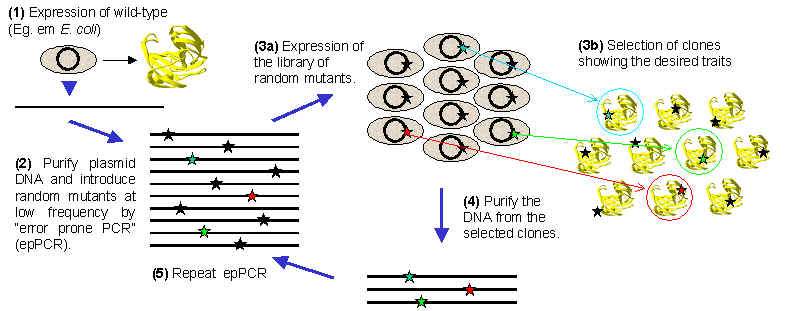
After the mutagenesis step is completed, the mixture of randomly mutated genes is screened for the selection of those mutants which show an enhancement of the desired property. The mixture of the randomly mutated DNA sequences is used to transform host cells, and after growing these transformed cells on a solid culture medium, individual colonies will appear (step 3a). Each colony is derived from a single transformed cell, and will contain copies of a single mutant from the total mixture. By selecting those cells which display an enhanced function for the protein of interest (step 3b), the mutation that is responsible for that change may be isolated and identified by nucleotide sequencing of the DNA (step 4). This clone selection process is crucial to the success of the directed evaluation strategy, and much care has to be taken at this point. At this point, the error-prone PCR may be repeated, followed again by steps 3a, 3b and 4, which will result in the accumulation of mutations in the target DAN sequence. Alternatively, the clones selected in step 4 can be fragmented and recombined as described below.
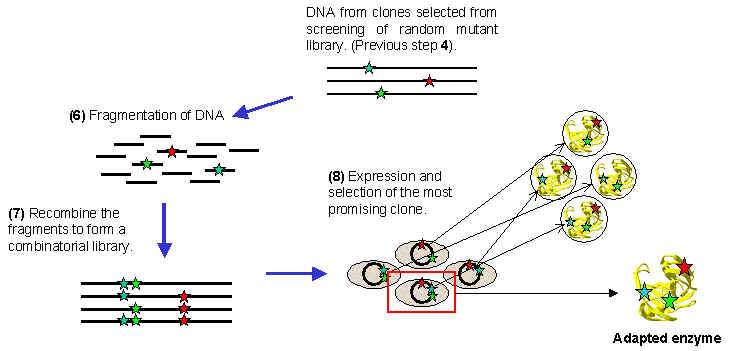
The mutants that are selected from the screening of the random mutant library enter into a process known as "DNA shuffling". In this technique, each of the DNA sequences that were isolated from the random mutant library are randomly fragmented (step 6), and the fragments are recombined in all the possible combinations to give what is called a "combinatorial library" (step 7), which will include all the possible combinations of the promising mutants that were obtained by screening the random mutant library obtained by error-prone PCR. Finally, screening of this combinatorial library (step 8) will identify a clone in which favourable mutants are combined in the best way to give the maximum improvement in the desired property of the adapted enzyme.