Xylanase 3D structure.
A wide variety of bacteria and fungi produce xylan-degrading enzymes, which they secrete into their immediate surroundings in order to break down the carbohydrate polymer xylan into shorter oligosaccharides which can then be used as an energy source by the microorganism. Xylanases have been grouped into families F10 and G11 on the basis of amino acid sequence similarity and three-dimensional structure analysis.
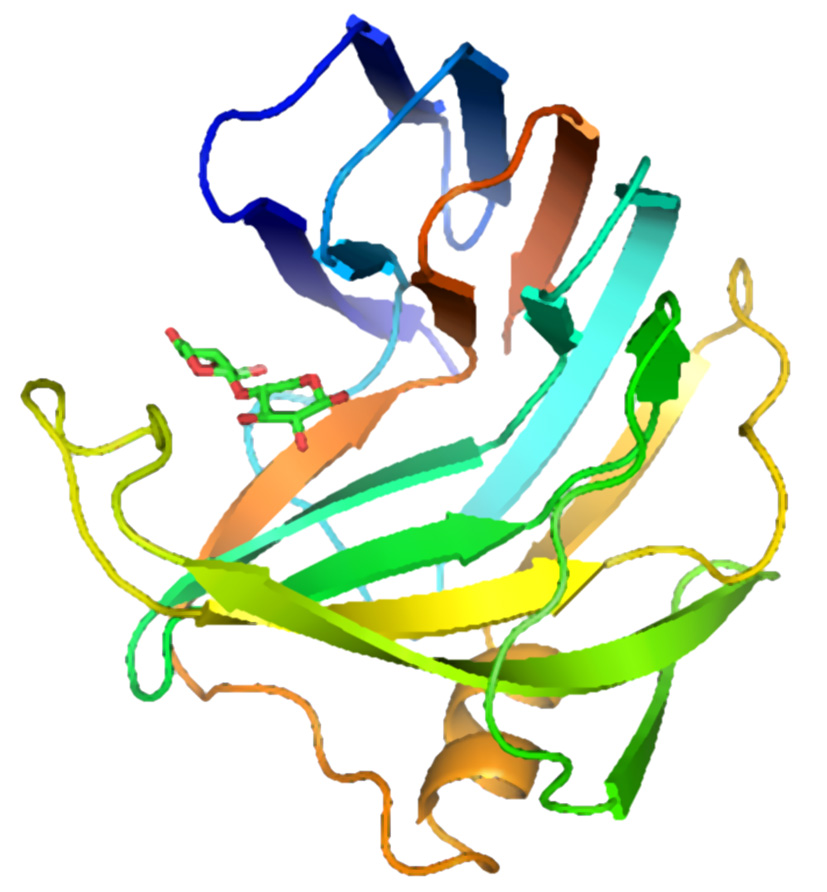
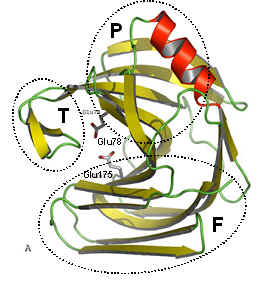
The figure above shows two views of a cartoon of the 3D-structure of the G11 xylanase from Bacillus subtilis (generally known as the xylanase A, or XynA) as determined by x-ray crystallography. The structure is typical of most G11 xylanases and, contains two twisted b-sheets forming a so called "jellyroll" fold which looks remarkably like a right hand, in which the individual b-strands thread back and forth to form the "finger" and "palm" domains (shown as the regions F and P in the diagram on the right). Residues lining the cleft formed between these two domains contribute to the substrate binding and active sites of the protein. An extended loop form the "thumb" domain (shown as T), which can open and close over the active site, so regulating the access of substrate to the catalytic region of the enzyme.